This is an overview of key takeaways from a recent paper publication, also presented as part of the TIA seminar series. This comprehensive study evaluated 20 pathology foundation models and determined that all 20 encode medical center information, leading to the potential for systematic diagnostic errors and clinical failures that prevent safe adoption. Researchers identified which models are more robust, demonstrated how spurious correlations cause misclassifications, and developed a robustification framework that helps improve robustness. The complete paper offers detailed methodologies, comprehensive analyses including slide-level studies and clinical impact assessments, and additional insights that provide a fuller understanding of the implications for the field. We encourage you to explore the full paper to gain the complete picture.
Background
Biomedical foundation models are becoming increasingly prevalent, and beginning to enter clinical validation and deployment. These models are trained with Self-Supervised Learning (SSL) using large, diverse datasets, which is often assumed to produce reliable general-purpose models. However, the challenge is that SSL captures any types of differences between samples including useful biological features, but also non-biological technical artifacts like variations in surgical techniques, tissue staining, or imaging equipment¹,². Ideally, models should differentiate between biological differences (ex. tissue type, cancer type) while ignoring confounding differences (ex. staining variations, equipment differences) that impact reliable generalization across different clinical settings.
This generalizability challenge is particularly important when evaluating data from different medical centers, where achieving similar performance is essential for reliable clinical usage. This study explored the effect of medical center signatures using PathoROB, the first pathology foundation model robustness benchmark built from real-world multi-center data. PathoROB features three novel metrics—the Robustness Index, Average Performance Drop, and Clustering Score—and uses four balanced, multi-class datasets covering 28 biological classes from 34 medical centers to measure how well models handle inter-institutional variations.
Robust models capture relevant biological features while remaining insensitive to confounding features. This overview focuses on the Robustness Index, a metric which quantifies whether biological features dominate over confounding features.
Methods
PathoROB Development
PathoROB was developed by subsampling four datasets from three public sources: Camelyon cohorts for tumor detection, two subsets from TCGA-UT for tumor type prediction, and Tolkach ESCA for tissue compartment classification. Each dataset was carefully balanced to ensure every medical center contributed the same number of cases, slides, and patches per biological class, enabling direct comparison between biological signals and medical center signatures.
Foundation Model Evaluation
Twenty publicly available foundation models with diverse training setups were evaluated, including various architectures, pre-training objectives, dataset sizes, and model capacities. This included Aignostics’ own Atlas foundation model co-developed with Mayo Clinic and Charité Berlin. Inference was run on each model as-is without fine-tuning, meaning images were processed through each model and the embedding outputs were used for analysis.
Each PathoROB evaluation began with balanced datasets but artificially introduced bias by adding more data from one hospital for specific classes, to test how bias affects downstream performance.
Robustification Framework
Since retraining foundation models is expensive, a comprehensive robustification framework was developed with mitigation techniques to improve robustness. These techniques included data normalization approaches like Reinhard and Macenko stain normalization, which digitally standardize stain colors to reduce variation. Additional methods included ComBat batch correction (originally developed for molecular data) and domain-adversarial training (DANN).
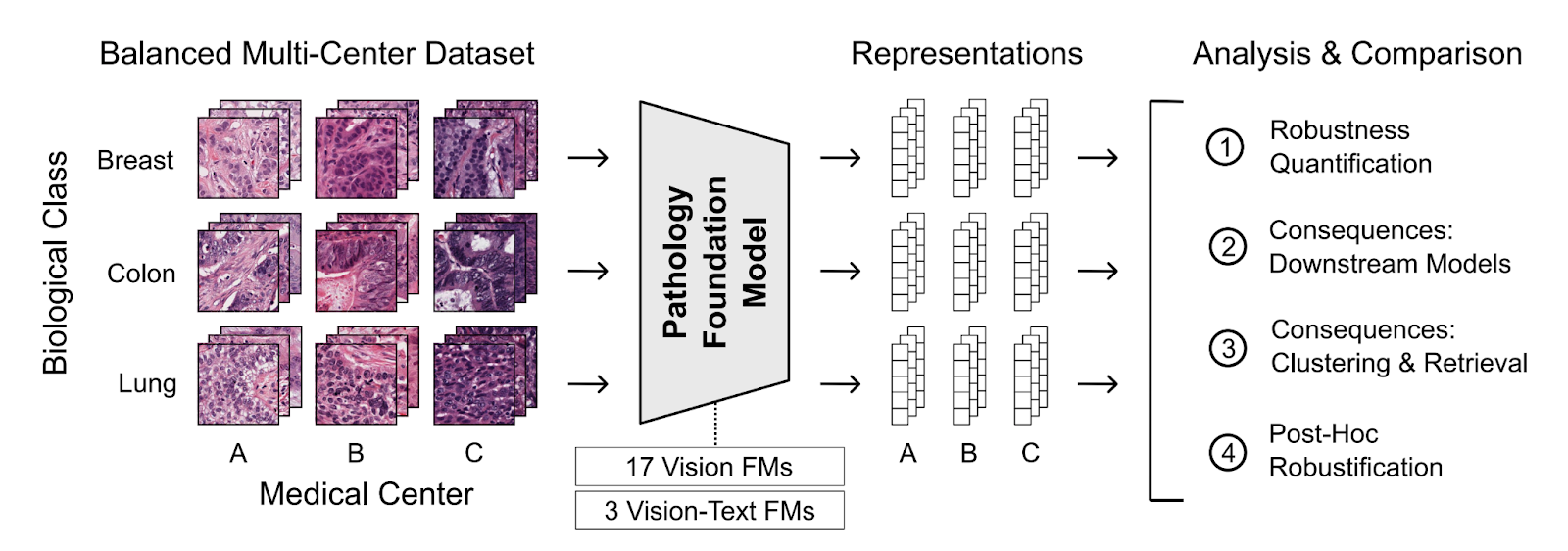
Results
Embedding Space Organization
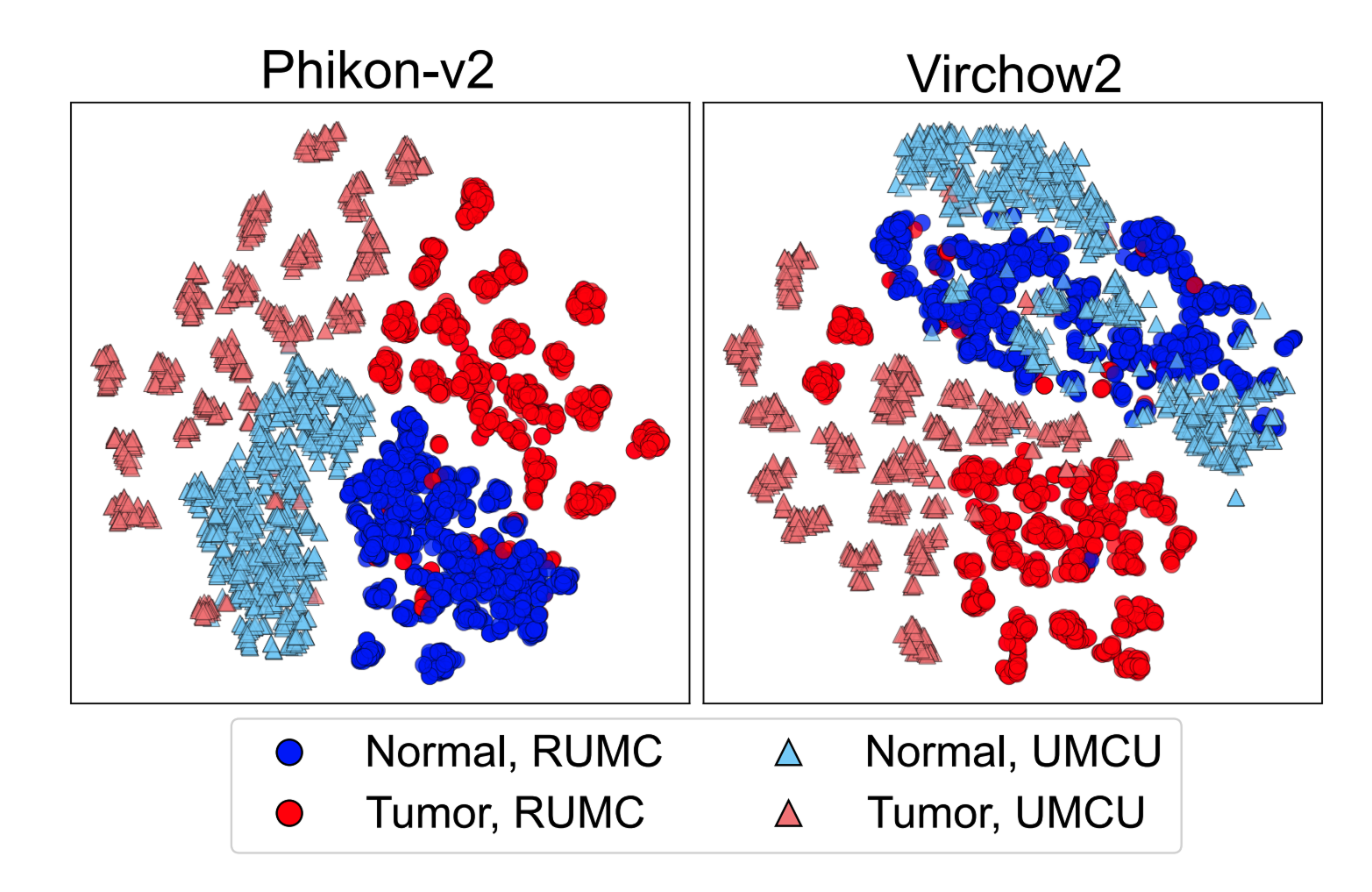
Starting with an exploration into feature space organization, medical center information was found to have a strong influence, as demonstrated through t-SNE plots (visualizations that place similar samples close together in two-dimensional space). The visualizations show clear differences between models: some like Phikon-v2 organized primarily by medical center (problematic), while others like Virchow2 organized mainly by biological information with secondary medical center grouping (better). Results varied widely between these patterns.
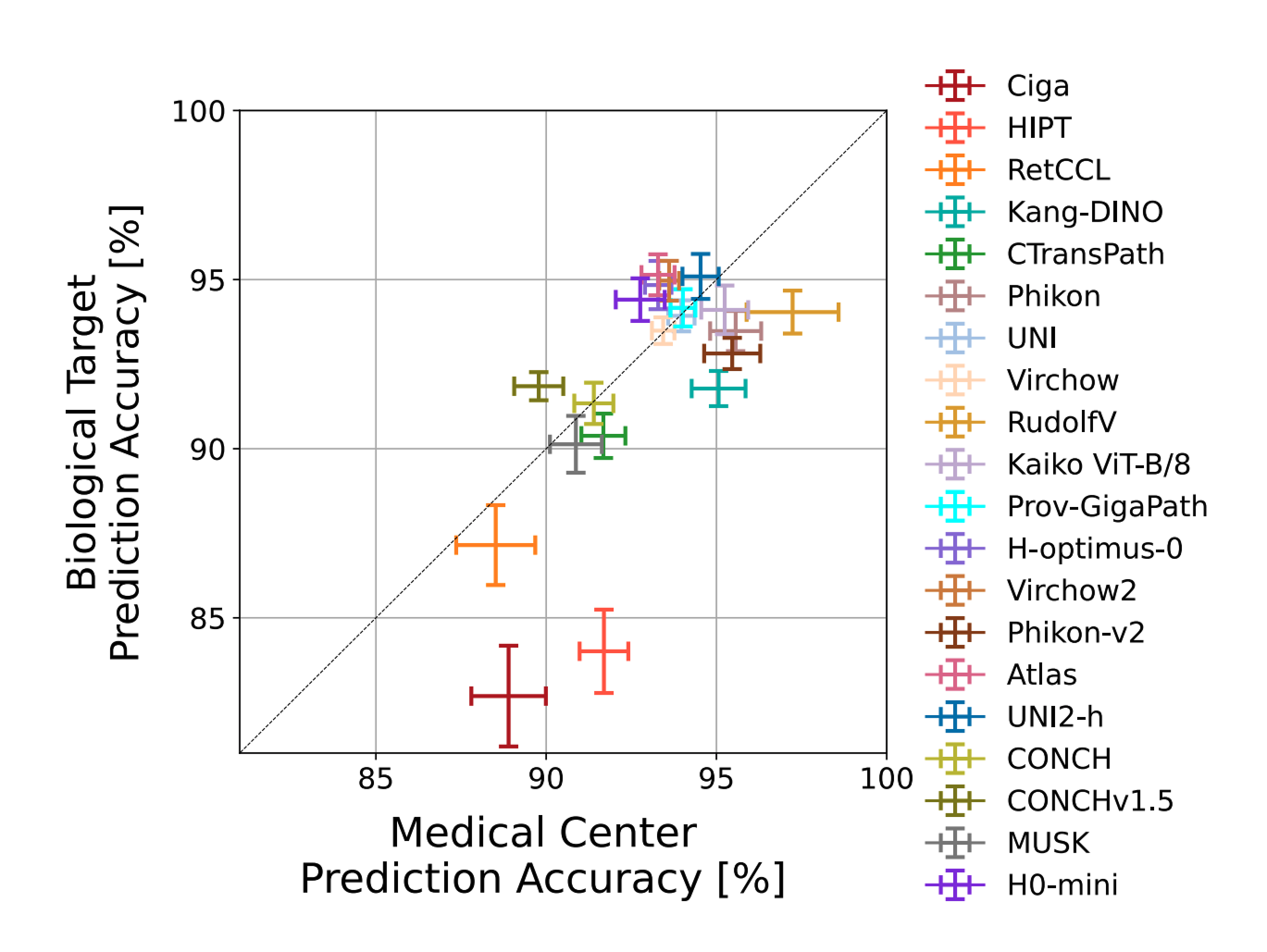
Feature Vector Analysis
Taking a deeper look at feature vectors, medical center origin was highly predictable with 88-98% accuracy across datasets. For more than half of the models, medical center prediction actually outperformed biological class prediction, suggesting that non-biological factors have a stronger influence on the representations than biological information which should ultimately drive medical decisions.
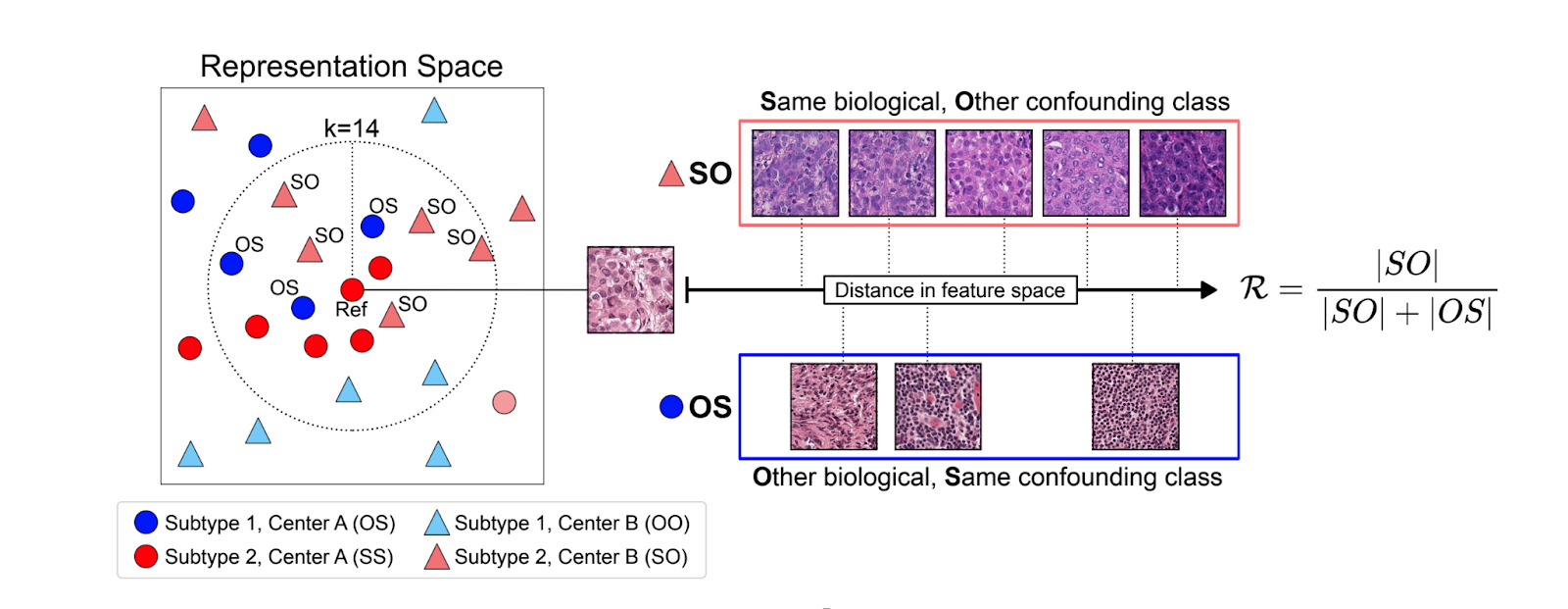
Robustness Index Results
The Robustness Index measures how much a model's embedding space prioritizes biological versus confounding features, ranging from 0 (not robust) to 1 (robust). For each reference sample, it examines neighbors that are either Same biological/Other confounding (SO) or Other biological/Same confounding (OS). Analysis revealed robustness scores ranging from 0.463-0.877 across models, with no model achieving full robustness.
Comparing robustness against training data size revealed a strong correlation (ρ = 0.692, p = 0.004) between the number of training slides and robustness. Larger datasets generally improve robustness, though some models exceeded expectations. Image/text models showed higher robustness than many vision-only models, likely benefiting from text information in medical reports.
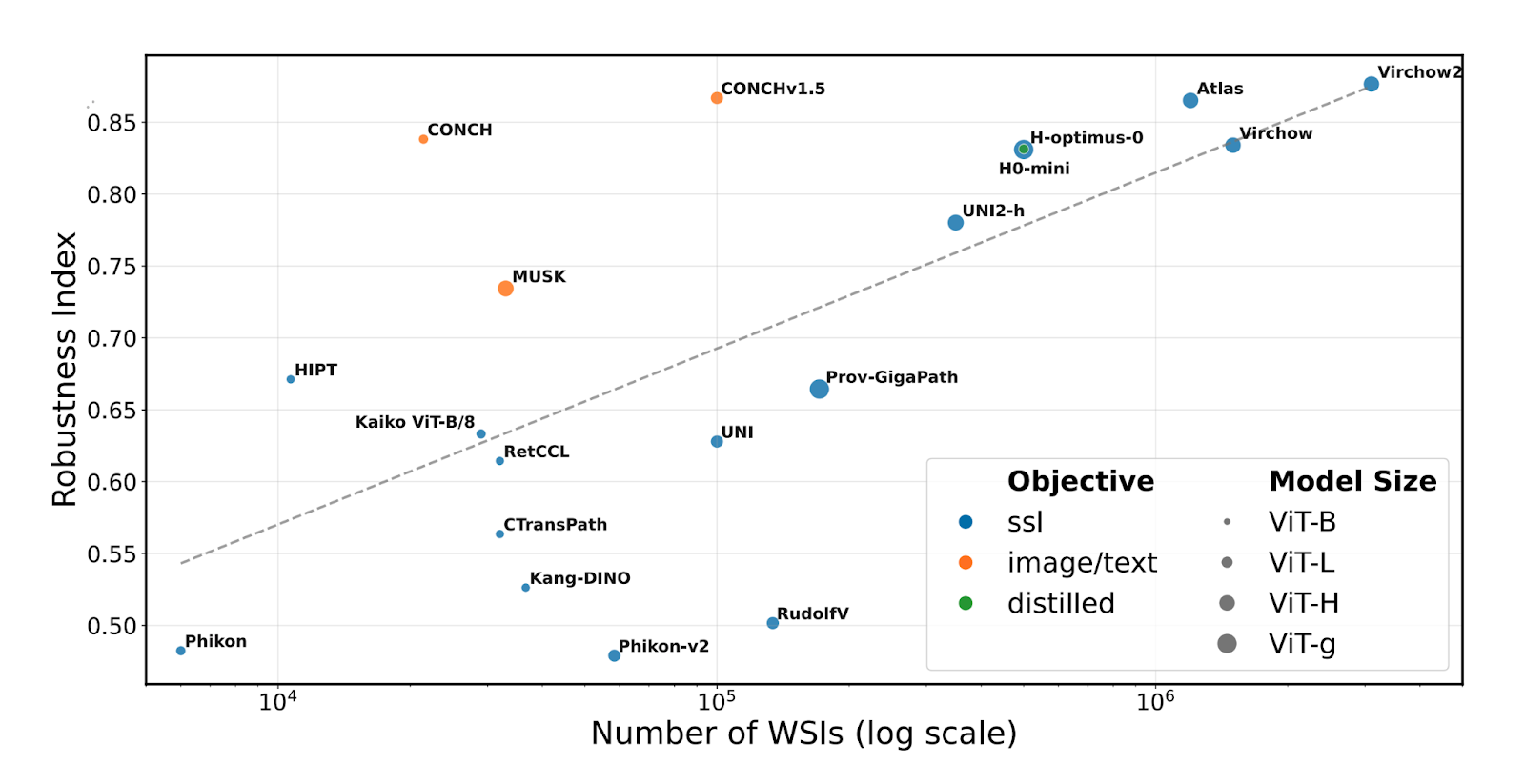
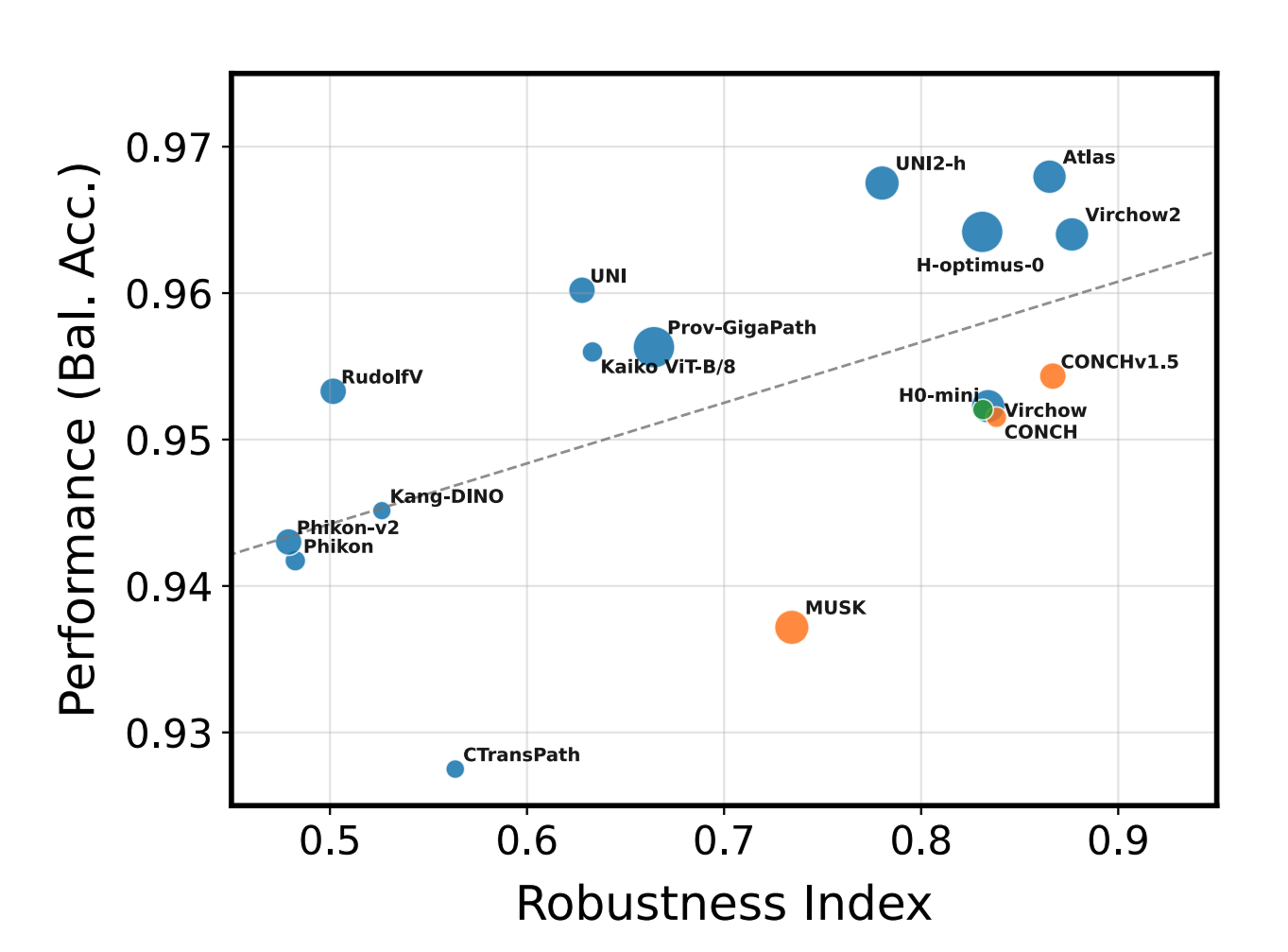
Within the examined models, our foundation model Atlas was among the models that provide the best tradeoff between accuracy and robustness.
Robustification Results
Testing the robustification framework showed promising but incomplete improvements. Data robustification (DR), in this case specifically Reinhard stain normalization, considerably improved robustness for most models with relative increases of +16.2% on average. Representation robustification (RR), in this case ComBat batch correction, enhanced robustness even further (+27.4% on average), with the most robust representations achieved by combining both methods (DR+RR).
However, no method completely eliminated performance drops, indicating that biological and technical information are often entangled, making complete separation challenging without risking removal of important biological signals.
Conclusion
This work demonstrated that while all 20 current pathology foundation models encode medical center information, some models show higher robustness than others. Notably, Atlas emerged as one of the two models achieving the best balance between accuracy and robustness. The robustification framework shows that some current limitations can be addressed, reducing diagnostic error risks.
The researchers hope this framework and benchmark will help others build better foundation models, advancing progress toward robust, clinically deployable AI systems that prioritize biological information over institutional artifacts. The complete paper contains extensive additional analyses including clinical impact studies, slide-level case studies, and comprehensive evaluations. We encourage readers to explore the full paper for complete results and methodology.
- Kömen, J., Marienwald, H., Dippel, J., & Hense, J. (2024). "Do Histopathological Foundation Models Eliminate Batch Effects? A Comparative Study." arXiv:2411.05489
- de Jong, E.D., Marcus, E., & Teuwen, J. (2025). "Current Pathology Foundation Models are unrobust to Medical Center Differences." arXiv:2501.18055

