Background
Developing accurate, scalable solutions for automated scoring of cell membrane expression from immunohistochemistry (IHC) slides will facilitate patient selection for clinical trials, better treatment decisions, and improved survival outcomes. Manual scoring methods, while still the gold standard, are inherently slow and suffer from inter-observer variability, creating challenges for consistency. Meanwhile, deep learning approaches offer impressive performance but operate as "black boxes"—they learn directly from data without providing insight into which image features drive their scoring decisions, making results more challenging to interpret.
To address this interpretability challenge, current automated approaches often attempt to provide transparency by using pathology annotations to train membrane segmentation models, and then calculating membrane expression from the segmentation result. While these methods provide transparency, they come with drawbacks: pathologists must provide time-consuming pixel-level annotations, essentially labeling each relevant pixel as membrane or non-membrane through careful boundary tracing, followed by complex post-processing steps.
This study aimed to demonstrate how using explainable AI (xAI) can capture the advantages of both interpretable and black-box methods. By requiring simple expression annotations that pathologists could assign at a glance, this method significantly reduced annotation burden while maintaining interpretability. Importantly, the xAI component allowed researchers to visualize which pixels influenced each prediction, enabling targeted identification of model errors and more precise training improvements.
Methods
Data was gathered from 48 non-small cell lung cancer (NSCLC) cases with PD-L1 staining, encompassing 78 whole slide images and 7,901 pathologist-labeled tumor cells. Pathologists first categorized cells across four expression levels: negative, weak, moderate, and strong. Then for model training, all positive classes (weak, moderate, strong) were grouped together, creating a binary classification task that predicted whether each patch was positive or negative for PD-L1 expression.
Model performance was validated using a hold-out dataset, achieving both accuracy and F1 scores of 0.97, demonstrating strong precision and recall. The team then applied Integrated Gradients, an open-source explainable AI technique, to generate heat maps highlighting the specific pixels that contributed to each prediction.
A key innovation of this approach is that no explicit cell detection or segmentation models were required. The classification model learned to identify cell boundaries and membranes autonomously, effectively creating pseudo-segmentation maps as a byproduct.
To validate model focus, the team computed optical density measurements—which quantify staining intensity through light absorption—with the expectation that highlighted pixels should correspond to areas of higher brown staining (indicating protein expression).
Results
Validation through optical density analysis confirmed the model's biological relevance, achieving a Pearson correlation of 0.87 between model predictions and pathologist annotations. This strong correlation demonstrates that the model focuses on clinically meaningful regions rather than spurious image features.
The explainable AI analysis revealed particularly compelling findings. Heatmaps showed that when making positive predictions, the model consistently isolated the membrane of the target cell without any explicit training on membrane annotations. As seen when filtering by xAI contributing pixels, rather than focusing indiscriminately on brown pixels throughout the patch, the model learned to distinguish between relevant staining on the target cell membrane and irrelevant staining from surrounding structures (Figure 1a).
Equally valuable was the ability to analyze model failures systematically. When errors occurred, the heat maps clearly illustrated the underlying causes: the model incorrectly focused on brown pigmentation from neighboring cell membranes or cells with darker nuclei rather than the target cell membrane (Figure 1b).
This level of insight would be difficult with traditional black-box approaches and provides actionable guidance for improving model training through targeted annotation strategies.
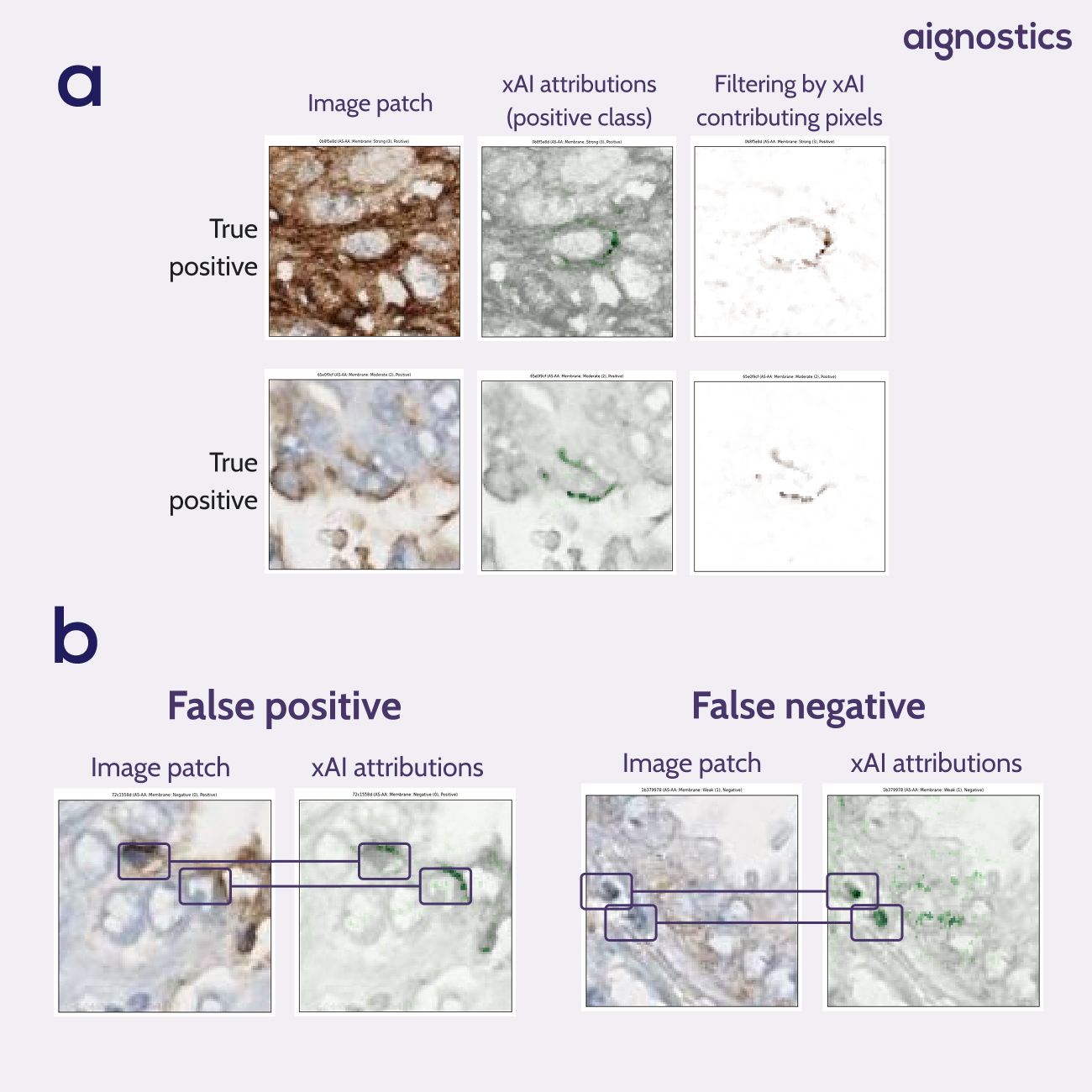
Fig. 1 xAI attribution heatmaps. a Heatmaps show the model has learned pseudo-membrane segmentation without having access to membrane annotations. Filtering on the contributing pixels shows a segmentation of positive membranes. b Heatmaps give insights into model errors. False positives show us that the model focuses on the membrane of neighboring cells. False negatives show us that the model ignores the center cell in favour of neighboring cells with a darker nucleus.
Conclusion
This study demonstrates that explainable AI can effectively bridge the gap between interpretable and high-performance approaches in medical image analysis. The approach's generalizability makes it particularly valuable—this xAI technique can be applied as post-processing to diverse medical imaging tasks beyond PD-L1 scoring. By using xAI, researchers can more effectively identify model shortcomings and explain mispredictions, allowing for more precise model training and improvement.

